Capstone Speaker
"Designing Visualizations to Enable Molecular Insights"
Jane Richardson (Duke University)
Abstract
 Our laboratory research goal of understanding 3D molecular structures
depends strongly on the development and use of visualization systems.
We especially value their effectiveness for enabling scientific insights,
in our original research process as well as in the communication of
those results, which leads us to emphasize interactive functionality
and pragmatic effectiveness over perfection as presentation images.
Therefore we design alternative molecular visualizations tuned to the
medium, the user, and the specific scientific problem at hand; feedback
from our own use improves them further.
Our laboratory research goal of understanding 3D molecular structures
depends strongly on the development and use of visualization systems.
We especially value their effectiveness for enabling scientific insights,
in our original research process as well as in the communication of
those results, which leads us to emphasize interactive functionality
and pragmatic effectiveness over perfection as presentation images.
Therefore we design alternative molecular visualizations tuned to the
medium, the user, and the specific scientific problem at hand; feedback
from our own use improves them further.
- Visualizations sometimes prompt scientific insight by suggesting metaphors from other realms, such as art. Examples influential in our work range from Greek vases to origami to Picasso.
- For students or colleagues approaching molecular structure from a verbal or algebraic pattern of thought, we work to develop their "3D molecular literacy". Best are interactive graphics exercises that let them explore, compare, and measure at their own pace, to master inherently 3D relationships such as handedness or hydrogen-bond geometry.
-
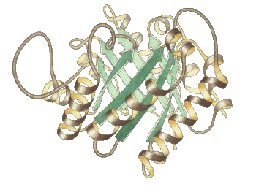 In our structure validation and improvement work, one challenge
is clearly visualizing the outliers from many quality measures
simultaneously on the molecular structure, to allow both rapid location
of 3D problem clusters and interactively-updated local detail as the
model is modified. Our current implementations support effective and
routine use in structural biology, but we seek further improvement of
their completeness and clarity.
In our structure validation and improvement work, one challenge
is clearly visualizing the outliers from many quality measures
simultaneously on the molecular structure, to allow both rapid location
of 3D problem clusters and interactively-updated local detail as the
model is modified. Our current implementations support effective and
routine use in structural biology, but we seek further improvement of
their completeness and clarity.
- A current quest is finding the right molecular problem and developing the measurement and analysis interface to show if an immersive virtual environment (the Duke DiVE) can provide superior scientific results over simpler systems. We plan to test interpreting the highly local experimental data that determine NMR structures.
- A recent visualization success that enabled new molecular insight is a simple but powerful way of interactively visualizing and clustering high-dimensional data. It enabled us to classify cleanly the noisy 7-dimensional dihedral-angle data that describes RNA backbone conformations, and to perceive the shortcomings of earlier attempts done either by automated clustering or by combining multiple 3D projections of the data.
Bio
Jane Richardson is a James B. Duke Professor of Biochemistry at Duke University. She and her husband David have worked together since the Sixties on research to determine, visualize, analyze, and understand the 3D structures of biological macromolecules. Professor Richardson pioneered ribbon drawings for representing protein structures, first drawn by hand and later adapted in almost every molecular graphics system. She first described many of the common features of overall protein folds and their local motifs (such as Greek key beta barrels, beta bulges, righthanded crossovers, helix caps, and now the subtle but influential backrub motion) and in recent years has diversified also to RNA, especially the details of its local backbone conformation. The Richardsons were among the early groups doing protein crystallography, helped start the field of protein de novo design, learned much from a 20-year collaboration with Fred Brooks in the UNC computer graphics lab, and in 1991 developed the interactive molecular graphics system of "kinemages". They have worked to spread molecular 3D literacy to students and colleagues at Duke and around the world. They developed a method that calculates hydrogen-atom contacts to visualize and quantify the details of packing interactions inside and between molecules, especially powerful for improving structural accuracy; it is implemented, with kinemage displays, in the MolProbity web service, which has supported more client worksessions than there are coordinate sets in the Protein Data Bank.
From a Swarthmore B.A. in Philosophy, Jane became a MacArthur Fellow, a member of the National Academy of Sciences and the American Academy of Arts and Sciences, a Biophysical Society National Lecturer and Phi Beta Kappa Visiting Lecturer, and a recipient (individually or with David) of the Emily Gray biophysical education award, Discover Magazine's Innovation award, and the Protein Society's Amgen Award. She now has three honorary degrees to fill in for her lack of a Ph.D.
 Week-at-a-Glance
Week-at-a-Glance